AWS Public Sector Blog
Genomic and Medical Big Data Go Serverless
Serverless architecture remains on the rise. From simple Alexa Skills to advanced web services, our customers’ use cases are innovative and diverse. Take Commonwealth Scientific and Industrial Research Organization (CSIRO)’s genomics research, for example.
“AWS has become versatile and interoperable making it possible to set up complex research workflows as serverless web services,” says CSIRO’s Dr. Denis Bauer. Dr. Bauer and her team created a bioinformatics research tool, GT-Scan2, which Jeff Barr featured two years ago as one of the first examples of serverless architecture for compute-intensive tasks.
Making sense of the human genome
Together with John Pearson’s Genome Informatics team from the QIMR Berghofer Medical Research Institute, Dr. Bauer’s team confronts one of the biggest challenges in bioinformatics – making sense of the human genome. They plan to use a serverless infrastructure, demonstrating its ability to handle both data and compute-intensive tasks.
“We have to look beyond research-scale solutions if we are going to realise the promise of genomics-driven precision medicine,” says Pearson.

Fig 1 Screenshot of the PhenGen-Insight tool showing a patient’s genetic risk for obesity and comparing his/her BMI against that of a peer group with similar risk.
What’s in your genes?
Genomic information is used often in clinical practice. Still, the genomic changes (variants) that cause diseases can be obscure. While better integration of genomics with clinical data from patient records will help show causation, the datasets are large and computing power immense.
The bigger challenge is the genomic information, which is frequently multiple gigabytes per subject and quickly scales to terabytes when thousands of subjects are involved. “Traditional database schemas don’t scale to cater for such large cohort sizes and are not economical enough to handle bursting workloads sustainably,” Aidan O’Brien from CSIRO said.
This challenge requires new ways to store, query, and link data. This is where serverless comes in.
“Working out which variants cause disease is not a needle-in-a-haystack problem, it’s a needle-in-a-bucket-of-needles problem,” says John Pearson, Genome Informatics Group manager at QIMR Berghofer and genomiQa co-founder.
Finding disease genes with serverless functions
The teams at CSIRO and QIMR Berghofer have joined forces to develop a prototype serverless genotype-phenotype (gen-phen) database on AWS. Here’s how the tool works:
1. Researchers and clinicians consolidate siloed datasets and filter in real time to identify medically similar patients. This is where the journey of piecing together the disease story begins.
2. The team then assigns a description to the clinical phenotype, or phenome. The Fast Healthcare Interoperability Resources (FHIR) standard and clinical terminology make this possible.
“Traditionally, Phen-Gen databases focus on either the medical or the genomics data and optimize accordingly, which limits the type of questions that can be asked,” Dr. Oscar Luo from CSIRO said. “By going serverless and embracing FHIR, we are bringing the two worlds together.”
3. Next comes implementation. An AWS Lambda function receives a query from the user, such as “all patients diagnosed with Melanoma under the age of 30,” and converts it into standard FHIR queries. They are then sent to the CSIRO FHIR server that contains the clinical data.
4. Finally, the medical and genomic information is merged client-side using AngularJS, which allows customized filtering to cater for the bespoke use case of each clinician.
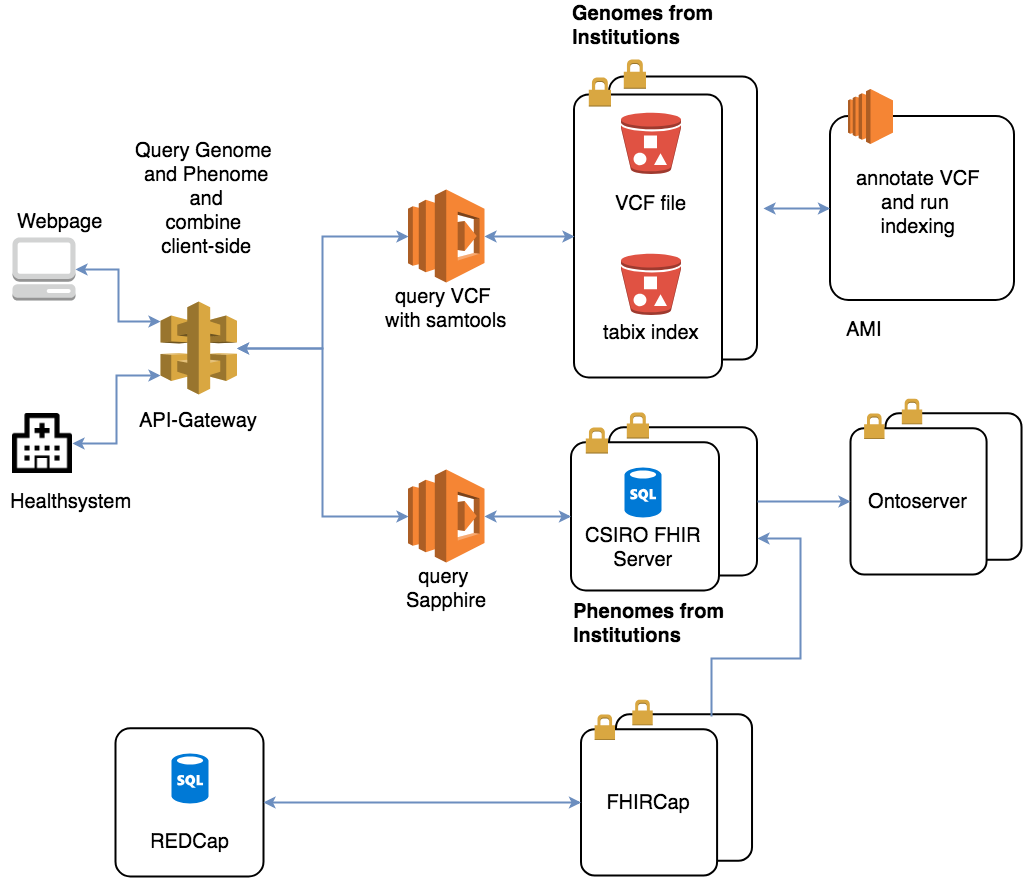
The team envisions the tool to scale to thousands of simultaneous users while remaining cost-effective. The solution does not pay for compute infrastructure to sit idle during periods of low or no use.
Benchmarking access time
“To arrive at the current optimal architecture we investigated several alternatives focused on speeding up the genomic file access,” says QIMR Berghofer’s Dr. Conrad Leonard. “While our current architecture is the fastest we could come up with for now, we identified new technologies that may allow for even faster solutions in the future.”
Amazon Athena is a promising candidate for the team. This AWS serverless query service allows real-time scalability to cater to the drastically varying traffic of clinical workflows. It can also perform complicated data transformations. At this stage, VCF files are converted to Parquet, which stores data efficiently in Amazon S3 and is accessible seamlessly through Athena queries. The conversion step makes use of technologies like Apache Spark and Hive. It is able to complete the one-off processing of several terabytes of data in minutes.
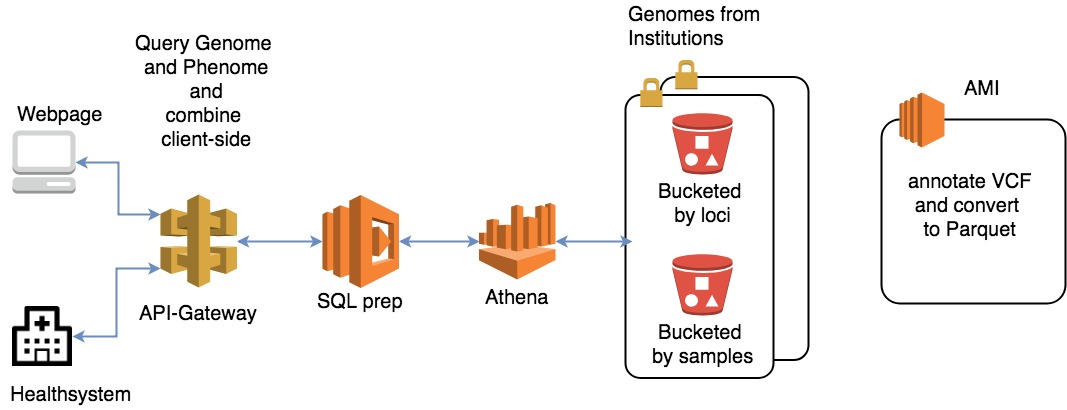
The team was able to measure the runtimes of these two setups using Epsagon, which allows workflow-based time measurements for serverless architecture.
How can a serverless Phen-Gen database impact clinical practice?
Put into practice, CSIRO’s Australian e-health Research Centre (AEHRC) and QIMR Berghofer use the system to deliver analysis and data-management capability for the Queensland Genomics Health Alliance (QGHA).
“The system avoids data silos while preserving data ownership and patient privacy,” Brian Thorne of CSIRO’s Confidential Computing Group said. “It does this by maintaining a separation between patients’ medical information and their genomic information, as well as isolating data across separate S3 buckets, which allows institutions to tightly control information release.”
“Innovations such as this are exactly the sort of technology that will support the implementation of genomics into clinical care to scale from the local to the national level,” adds AEHRC’s CEO, Dr. David Hansen.